Crossing paths with bugs, issues, unknown systems, and more
25 May 2024From my experience working at Simpl.
In this article, I jot down, different issues & bugs I or my teammates faced in software systems along with fixes we did. Note that a lot of them are quick fixes which we did in the interest of time and escalation. Long term fixes for them were implemented as well - that part, will probably cover in another blog. I may miss a few of them, or rather its just tip of the iceberg, but have added as many as my memory serves me right. I think a lot of my learnings actually came from solving issues.
- Issues with data pipeline(s):
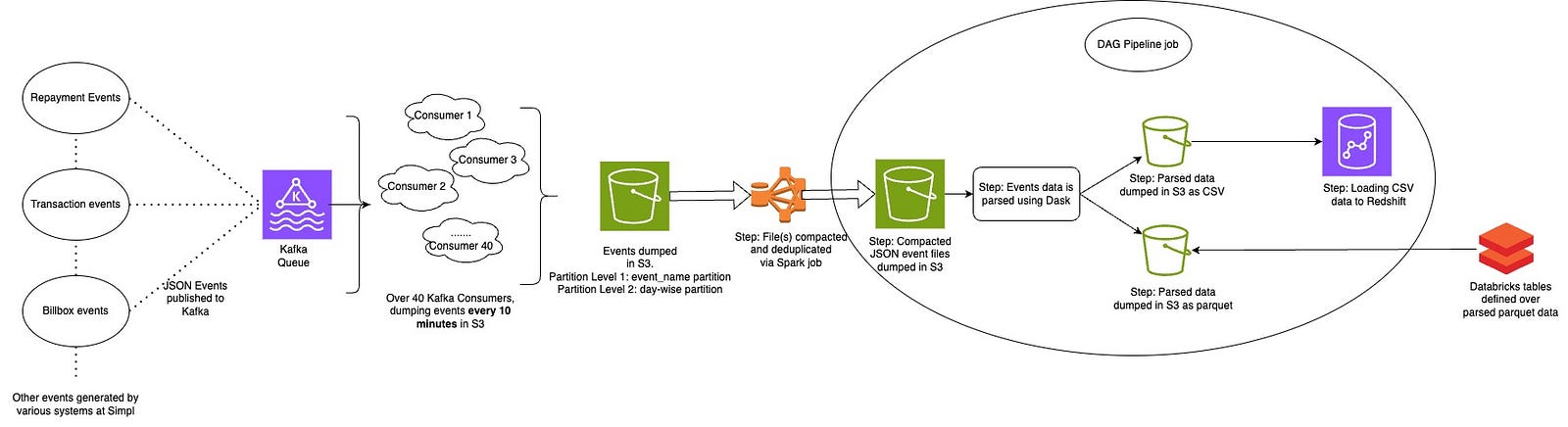
- Pipeline 1: As seen below, at a high level it’s an event parsing pipeline. Kafka consumers dump data in S3. Since the frequency of dump is high, large number of small sized files are created in near-real-time. While there are other nrt table(s) dependent on these files, for our case, we later compact & dedup these files using a spark job and rewrite them in a path. After compaction, parsing related workflows run to parse the data and dump it in different locations in parquet and CSV format. External tables from Databricks are created on top of parquet files, and the CSV files are used to load in Redshift tables. Redshift/Databricks tables cater different needs of downstream teams.
- Issue fix 1: CSV data to Redshift load job failure due to integer overflow value (Integer valid range -2147483648 to 2147483647) for one of the records in CSV. More details were found in Redshift’s inbuilt table:
stl_load_errors. It was a corrupt data found when deep investigating, removed the record in raw json and re-loaded the flow. We also added a key in the Redshift data load COPY command similar toIGNOREALLERRORS, though not ignoring all, we had a limit added, if it would breach it, we would be notified something has failed to load, if its few records, then ignore errors and load anyways. Another issue of similar flavour was observed in later days which was null values in records in CSV: event_timestamp of the null record row: c423b113-5d05-4ae0-a659-8d6ea42ea3e8,,,,,,,,,,,,,,,{}. Similar fix had to be followed. - Issue fix 2: Data parsing from Dask failed due to issues around dask schedular not being able to communicate with dask workers. Redeployment of the ecosystem and rerunning the job fixed it. Had it failed again, I would have checked deeper into root cause.
- Issue fix 3: VPC endpoint changes made for some systems by infra team, impacted DE workflows. Debugged and fixed issue with them.
- Issue fix 4: Spike in one of the events caused issues parsing it. Temp fix was increasing the number of partitions in df which is used in parsing. Types of errors observed:
distributed.comm.core.CommClosedError: in <TCP (closed) Scheduler Broadcast local=tcp://10.88.13.127:47998 remote=tcp://10.88.12.63:39843>: Stream is closed;)orOSError: Timed out during handshake while connecting to tcp://10.88.12.115:34253 after 30 s;) -
Issue fix 5: Cases where we were getting malformed JSON from Kafka consumers for some records causing parsing issues downstream. The faulty records were pin-pointedly identified from below pseudocode, and removed from raw json and workflow was rerun.
We had to pinpointedly identify few things: Which event was causing the issue (since a parser may parse multiple events) Inside the event, which exact JSON.gz file was causing JSON issue (as it wouldn’t usually be that all the JSON files we get from Kafka have issues) Inside the JSON.gz file, which exact line/record did we see the malformed JSON data which was not getting JSON parsed. for f in files: path = "s3://" + f # Adding s3:// tag... try: _log.info(f'Read file: {path}') import dask.dataframe as dd df = dd.read_json(path, storage_options=_get_s3_creds()).compute() # trying to read each JSON in directory in dask, if we are unable to read it, means, there is error in JSON file except: print(f'Unable to read file, as errors might be present in data for gzip file: {path}') error_gzip_files.append(path) _log.info(f"Error causing gzip files are: {error_gzip_files}") return error_gzip_files with gzip.open(obj, "r") as f: _log.info(f'Reading each line of JSON data in the gzip file to find the corrupted record') for row in f: try: row = json.loads(row) _log.info(f'Parsed JSON line number: {json_record_file_line}, in the JSON gzip file') except: _log.info(f'Error found while parsing JSON record in line number: {json_record_file_line} present in data for the gzip file') json_record_error_line_list.append(json_record_file_line) json_record_file_line = json_record_file_line + 1 - Issue fix 6: Similar to the data spike issue discussed earlier, so the system parses different events in parallel in Airflow tasks. We had a similar spike case for one of the heaviest events, this time, we let the heaviest parser complete the parsing first utilizing all resources of cluster, after that all the smaller parsers were run in parallel increasing airflow task pool size.
- Issue fix 7: There was failure in compaction job in morning - S3 delete (which is used in compaction code) was throwing some error probably due to too many requests. Failure led to too many files of small size getting created, and parsing job was failing due to small files problem, hence we fixed and reran compaction job, I recall fix was decreasing the rate at which we delete files, once files were compacted, we reran parser jobs.
- Issue fix 8: Airflow pool size for set of tasks was high, thereby it was scheduling lots of jobs - which indeed started to fail as distributed cluster was not able to handle it. We later reduced airflow pool size.
-
Issue fix 9: Airflow infra resides in EC2 machines, any change in related code is deployed to EC2 machines. Deployment involves pip install steps. There were issues around some packages having conflicting versions on prod. It was fixed by fixing dependencies manually in machine and later updating the requirements txt file used for deployment. A better fix would be to dockerize the service as well as use something lke poetry to anchor package versions.
ssh ubuntu@10.x.x.x jsonschema --version sudo pip install jsonschema==3.2.0 --force-reinstall sudo pip install setuptools==71.1.0 --force-reinstall sudo systemctl restart airflow_scheduler # could also use systemctl stop/start commands sudo systemctl restart airflow_webserver sudo systemctl restart airflow_worker sudo systemctl status airflow_scheduler sudo systemctl status airflow_webserver sudo systemctl status airflow_worker # To figure out mismatched packages, we compared packages in staging and prod machine and fixed it... pip freeze > requirements_staging.txt exit - Issue fix 10: Had a requirement to re-parse old data (of some years) for some events in table. Did an interesting thing, created a temporary backfill workflow and pointed staging to prod tables and (same temporary workflow in prod) prod workflows anyways were inline. And triggered backfill workflows of both envs to prod, which ran non-stop and was loaded; When our other critical workflows were triggered on prod, since our machines were already running heavy with backfill, they did not have resources and when other workflows too got triggered, the over burden killed the machines and there was outage. Had to fix things, and slow down the backfill in chunks and ensuring when critical workflows run, backfill is not running. Of course, this could’ve been handled in better way.
- Issue fix 11: Had a case where the Airflow PythonOperator task was not recognising the datetime library, this happened because of the new Python version we had upgraded to, which involved using Pendulum library rather so fixed using the same.
- Issue fix 12: All task/job logs in Airflow UI disappeared, and were not visible - both current day and earlier day, it was caused due to tinkering/putting bad value in Airflow connections list in the UI. Fixing value there fixed the issue - it was related to a variable having credentials to connect to S3 where logs are persisted.
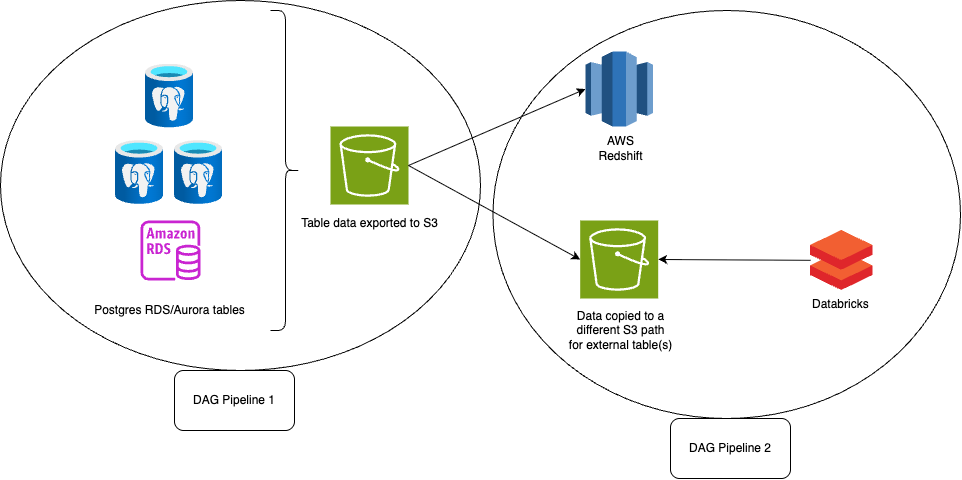
- Pipeline 2: As seen below, at a high level it’s a snapshot batch job pipeline where snapshot/backup of the OLTP Postgres tables is taken everyday and exported to S3, data of it is accessed from different OLAP tables. As running analytical queries on OLTP tables can be heavy. Basically, there is a snapshot scheduled at AWS RDS end for tables everyday. Once the snapshot is ready, there are workflows which export this data.
- Issue fix 1: There were issues around Redshift load when length of a attribute or column in data record was longer than length defined in table where it was to be loaded or different datatypes in file vs dtype defined in table column, etc. More details were found in Redshift’s inbuilt table:
SVL_S3LOG. - Issue fix 2: RDS export requires unique naming for every export. If same name is used - it will yield error:
Error Occurred while starting export task | ExportTaskAlreadyExists. If for some reason, the export job triggered from Airflow reruns, then issue would stem from it. Although, once export is triggered in RDS, its independent, but still issues may come up. These were fixed by changing name in format and rerunning export:. I think could be better if format had been: - Issue fix 3: Upstream tables in RDS were deprecated, hence when they were exported by the system there was no data causing impact in some of the workflows. Dependency from them was removed.
-
Issue fix 4: RDS Redshift tables are loaded from parquet data. If there is a mismatch in columns in underlying parquet and the defined table, the new columns are dropped or created with null value + datatype typecasting is done, if no mismatch then data loaded as it is -> Code is designed in such way. In one case, there was a mismatch case, but even after typecasting, loading it to Redshift was giving issue, hence in the end had to create the missing columns so its loaded directly.
# Sample pseudocode of typecasting for field in agg_data_table.fields: col = field.name if col not in raw_df: missing_cols.append(col) raw_df[col] = None df[col] = raw_df[col].fillna(get_default(field)).astype(get_type(field)) def get_type(field): if isinstance(field, Timestamp) --> return 'datetime64[us]' elif isinstance(field, Integer) --> return 'int32' elif isinstance(field, BigInt) --> return int elif isinstance(field, Float) --> return float elif isinstance(field, Boolean) --> return bool elif isinstance(field, Varchar) --> return str elif isinstance(field, JSONVarchar) --> return str default --> return str def get_default(field): if isinstance(field, Timestamp) --> return 'NaT' elif isinstance(field, Integer) --> return 0 elif isinstance(field, BigInt) --> return 0 elif isinstance(field, Float) --> return 0 elif isinstance(field, Boolean) --> return False elif isinstance(field, Varchar) --> return '' elif isinstance(field, JSONVarchar) --> return '' default --> return ''-
Issue fix 5: Received issue in workflow:
An error occurred (ExportTaskLimitReachedFault) when calling the StartExportTask operation: You have reached the limit of 5 concurrent export tasks). In short, correction was in the way snapshot export code was defined. Different db’s were exported from the RDS cluster in the code. It was defined earlier that for each db export it will initiate an RDS cluster export of which the db is a part of, this caused multiple times export being initiated and raising error from AWS end. Correction was, initiate RDS cluster export once, and export all related db’s in it in a single go. RDS snapshot → Specifies the db(s) once export is complete → Later dumps the selected db’s table(s) parquet files to S3.# Code: Observe have to pass multiple db's list response = client.start_export_task( ExportTaskIdentifier=task_name, SourceArn=source_arn, S3BucketName=s3_bucket, IamRoleArn=iam_role, KmsKeyId=kms, S3Prefix=s3_prefix, ExportOnly=[f'db1', 'db2', 'db3', ...etc] ) -
Issue fix 6: Similar to the flow discussed in this section, we have a flow where we restore the RDS data to another RDS cluster instance. Later there are workflows querying on the restored instance. Issue observed was poor query writing and incresing data made the cluster further heavier, bad query was bringing down the db. Optimized query and increased capacity. Also, had explored option to use something like pandas sql chunk size option, but it seems it retrieves whole dataset from db and then chunks the response and returns, rather than working on some query level optimization and chunking - so did not use it.
Old query: Joining first and then filtering select u.phone_number, max(v.limit) as limit from table1 u inner join table2 v on v.user_id = u.id where v.approver = 'ABCD' and u.phone_number in ('9999999999', '9999999990',... etc) group by u.phone_number, v.approver New query: Filtering first then joining select u.phone_number, max(v.limit) as limit from ( select * from table1 where phone_number in ('9999999999', '9999999990',... etc) ) as u inner join table2 v on v.user_id = u.id where v.approver = 'ABCD' group by u.phone_number, v.approver
-
- Other pipelines:
- Issue fix 1: In one pipeline, we had a sqoop workflow in an EC2 machine, which fetched data from external db’s to the machine, and dumped it to S3, meanwhile it would clean-up the in-memory data present after that. Sometimes, the data cleanup would not happen properly bringing down the machine, this was checked and fixed.
- Issue fix 2: We have deployment pipelines built using Concourse. While setting up deployment pipeline for a service - wherein in short, code was deployed to EC2 machines using ansible. There were some secrets stored in ansible vault which had expired. Had to decrypt, change the key, encrypt back the vault and fix. There were some other issues as well, which I have captured in another blog.
-
Issue fix 3: Had a flow where data which is about to be loaded to a table is enriched with some zipped mmdb data. Zip had multiple files and mmdb data. One time, there was some corruption in data, had to manually fix it and rerun flow.
(base) ➜ gunzip file.tar.gz ## unzipped the tar file using gunzip to ensure any metadata doesn't get corrupt (base) ➜ tar -xvf file.tar ## then expanded the files inside using tar command, We could also use tar -xzvf <> command to unzip the file, but in earlier attempt it was causing problems reading the files later on when used for enrichment logic, so better use gunzip to unzip it first ## --- multiple files including data.mmdb, LICENSE.txt, COPYRIGHT.txt, etc. (base) ➜ tar -cvf gunzipTarGeoIP2-City.tar GeoIP2-City_20230512/ ## Converting back (base) ➜ gzip gunzipTarGeoIP2-City.tar ## Used gunzip to zip back tar file -
Issue fix 4: Some pipelines were running queries on Redshift to access/load data. Due to some heavy queries, Redshift got deadlocked. We had to kill queries to release resources (later reran them separately)
## To get details around locking queries in Redshift: select * from pg_locks select * from STV_SESSIONS
- Pipeline 1: As seen below, at a high level it’s an event parsing pipeline. Kafka consumers dump data in S3. Since the frequency of dump is high, large number of small sized files are created in near-real-time. While there are other nrt table(s) dependent on these files, for our case, we later compact & dedup these files using a spark job and rewrite them in a path. After compaction, parsing related workflows run to parse the data and dump it in different locations in parquet and CSV format. External tables from Databricks are created on top of parquet files, and the CSV files are used to load in Redshift tables. Redshift/Databricks tables cater different needs of downstream teams.